in vitro PARP Assay
DNA Damage Response (DDR)
DNA damage occurs in mammalian cells in many different formats, including single-strand break (SSB), base oxidation, double-strand break (DSB), DNA adductions, and intra-strand and inter-strand crosslinks [1]. Different types of damages elicit different molecular and cellular responses, which together form an integrated signaling network that plays a crucial role in the surveillance of DNA integrity and maintenance of genome stability.
Effective repair of DNA damage imposes significant pressure, particularly, to highly proliferative cells including cancer cells. Most cancer cells appear to be impaired in their DDR capacity and become more sensitive to the imposed DNA damage burden than normal cells.
Much efforts have been put into looking for drugs that inhibit the DDR, as this is expected to enhance the effectiveness of radiotherapy and chemotherapies for cancers. For example, small-molecule inhibitors of key DDR components such as topoisomerases and other nuclear enzymes have been developed and tested in many cancer clinical trials [3]. To support drug discovery in this field, WuXi Biology offers multiple assays to screen and characterize topoisomerase inhibitors.
Gel-based assay of topoisomerase I
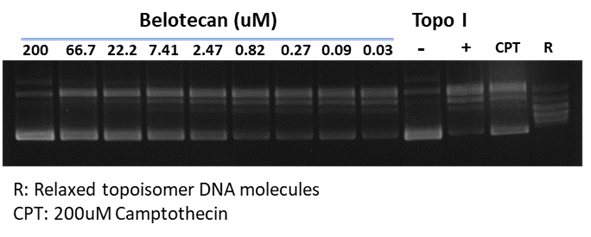
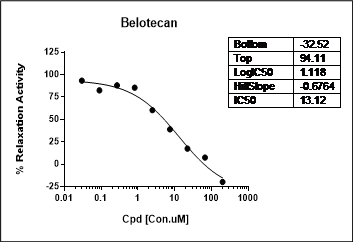
Certain DDR inhibitors, such as the check-point kinase CHEK1/CHEK2 (aka CHK1/CHK2) inhibitors, may block apoptosis and thus alleviate drug toxicity in normal cells. These two Ser/Thr kinases, by regulating p53 and cyclin dependent kinase CDK2, represent a key element of the homologous recombination mechanism of DDR. WuXi Biology has developed assays for these targets, including CHEK1/CHEK2, CDKs, WEE1 G2 checkpoint kinase, and the p53 regulator MDM2.
Different DDR pathways have overlapping/complementary functions. Since tumor cells are often impaired or lack certain DDR capacity, this has raised the possibility to elicit synthetic lethality by inhibiting complementary pathway(s) that would otherwise thwart the imposed selective pressure. This has been demonstrated in the case of PARP inhibition, which produces strong, selective effects against BRAC1/2 mutant tumor cells that are defective in homologous recombination. Discovery of novel PARP inhibitors is of great interest to promote PARPi therapies [4].
Poly (ADP-ribose) Polymerase (PARP) Biochemical Assays
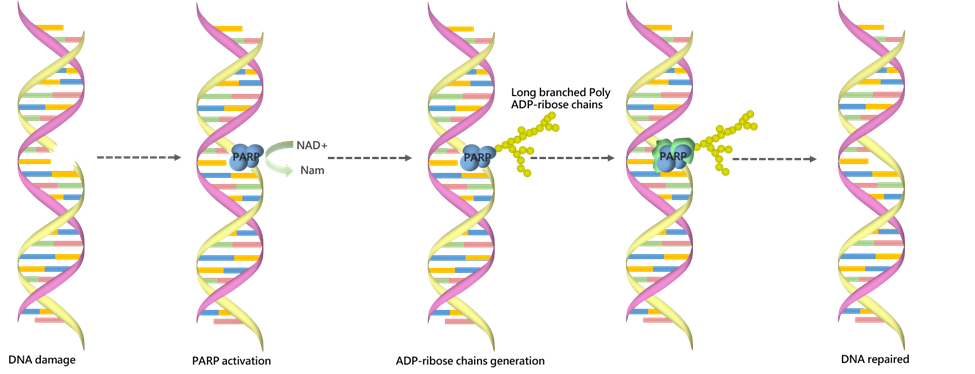
Poly (ADP-ribose) polymerase is a family of enzymes that play an important role in many cellular processes such as DNA repair, cell death, chromatin functions and genomic stability [5]. PARPs can transfer ADP-ribose from NAD+ onto target nuclear proteins forming long branched poly ADP-ribose chains. WuXi Biology offers a panel of PARP-related biochemical assays to support customers’ discovery needs.
Compound screening options in both low and high-throughput formats are available. Customized assay development can also be provided, as multiple PARPs can be expressed and purified in-house by our protein production team.
PARP1 assay
Among a family of structurally-related ADP-ribosyltransferases (ARTs), only the first six members are thought to be bona fide PARPs that catalyze cleaving of NAD+ and transfer the ADP-ribose moiety to their target proteins [6]. Of these, the founding member PARP1 has attracted strong interest for drug discovery for two reasons: 1) monotherapy based on PARP1 inhibitor was first shown to preferentially kill tumors that harbor the BRCA1 and BRCA2 mutations, 2) PARP inhibitors elicit cytotoxicity in the presence of chemotherapies that induce single-strand DNA break [7]. To support discovery of novel PARP1 inhibitors, we offer the following related assays.
– PARP1 Chemiluminescent Assay (96/384-well plate)
This ELISA-based assay consists of three major steps:
- The plate wells are coated with histone proteins.
- Biotinylated NAD+, PARP1 enzyme and activated DNA are added into the wells and incubated together, which allow the incorporation of biotinylated poly ADP-ribose chain into the histones.
- Wells are incubated with streptavidin-HRP followed by addition of an ECL substrate for detection.
Case study of PARP inhibitors
– PARP1 Trapping Assay (96/384-well plate)
In this fluorescence polarization (FP) assay, PARP1 binds to a fluorophore-labeled oligonucleotide duplex probe and forms a stable complex, which produces highly polarized light upon excitation. Upon NAD+ substrate addition and enzyme activation, PARP1 dissociates from the oligonucleotide duplex and rotates freely, thus reducing emission of polarized light. A PARP1 inhibitor is expected to cause trapping of PARP1 to the DNA, and hence increases the FP signal in a dose-dependent manner.
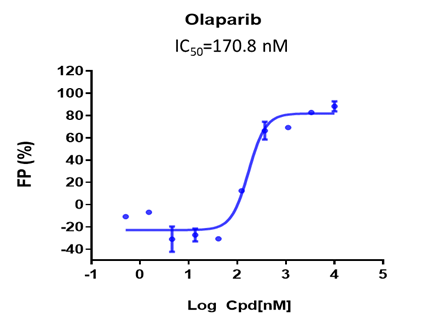
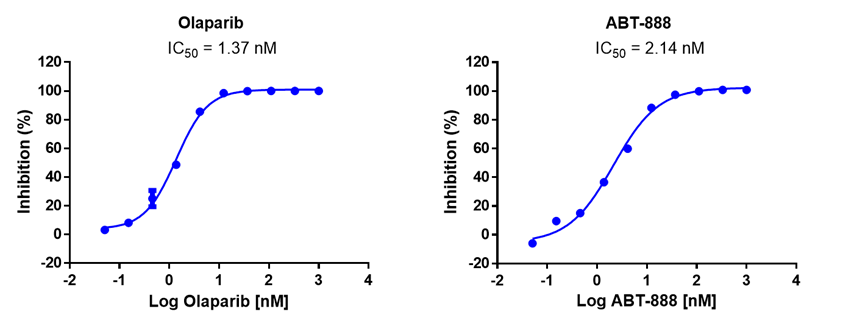
Case study of PARP inhibitor Olaparib
PARP2 Assay
Similar to PARP1, PARP2 also contains a catalytic domain and catalyzes the NAD+-dependent poly(ADP-ribosyl)ation; yet unlike PARP1, PARP2 lacks the zinc-finger DNA-binding motifs that are present in the N-terminal domain of PAPR1. Many PARP1 inhibitors also suppress enzymatic activity of PAPR2.
– PARP2 Chemiluminescent Assay (96/384-well plate)
This assay also takes an ELISA-based approach, similar to that used in the analysis of PARP1.
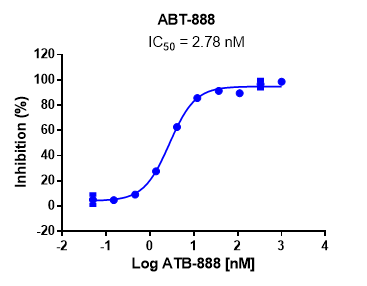
Case study of PARP inhibitor ABT-888
– PARP2 Homogeneous Assay (384-well plate)
This assay utilizes the AlphaLISA platform, which relies on proximity-based energy transfer through oxygen channeling. Specifically, PARP2 is incubated with activated DNA and biotinylated substrate, and then acceptor beads, followed by incubation with primary antibody and donor beads.
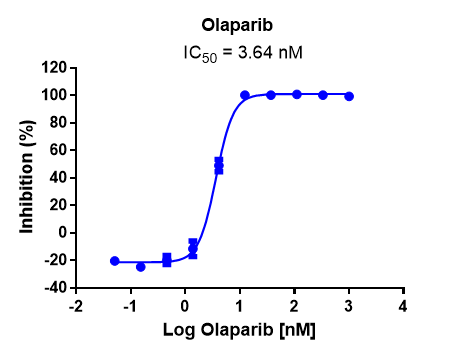
Case study of PARP inhibitor Olaparib
Other PARP-related assays
WuXi Biology has the capability to set up experiments for other PARP proteins, and PARP-related assays can be customized according to client’s needs.
Prolonged incubation of PARP inhibitors are known to increase DNA damages in certain tumor cell lines, as suggested by the accumulation of DNA damage-associated markers (such as g-H2AX and 53BP1). We have developed imaging analysis of these markers. Moreover, we have established an analysis pipeline of DNA damage markers in multiple cancer cell lines upon irradiation or drug treatment, and these platforms also support high-content screening (HCS) format.
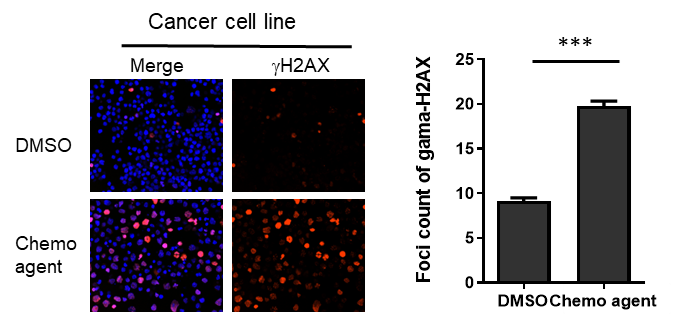
References:
[1] Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009 Oct 22;461(7267):1071-8. doi: 10.1038/nature08467.
[2] Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127-33. doi: 10.1101/sqb.2000.65.127.
[3] See https://clinicaltrials.gov/
[4] Rose M , Burgess JT, O’Byrne K, Richard DJ, Bolderson E. PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance. Front Cell Dev Biol. 2020 Sep 9;8:564601. doi: 10.3389/fcell.2020.564601.
[5] Herceg Z, Wang ZQ. Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat Res. 2001;477(1-2):97-110. doi:10.1016/s0027-5107(01)00111-7.
[6] Steffen JD, Brody JR, Armen RS, Pascal JM. Structural Implications for Selective Targeting of PARPs. Front Oncol. 2013 Dec 20;3:301. doi: 10.3389/fonc.2013.00301.
[7] Chen A. PARP inhibitors: its role in treatment of cancer. Chin J Cancer. 2011 Jul;30(7):463-71. doi: 10.5732/cjc.011.10111.

