Molecular Glues Discovery: Challenges and Opportunities
Introduction:
Molecular glues (MGs) are a class of small molecules that alter the surface properties of proteins to strengthen or induce protein-protein interactions (PPI), leading to specific biological effects like protein degradation, pathway inhibition, or activation. Their unique properties endow them with high selectivity and bioavailability, allowing for controlled drug release. Molecular glues show great clinical potential in treating diseases such as diabetes, cardiovascular diseases, ophthalmic disorders, and cancer.
Molecular Glues and Targeted Protein Degradation
Most diseases arise from disruptions of the normal interactions between proteins within signaling pathways. Manipulation of these interactions is a novel promising strategy for regulating protein biological functions and developing disease treatments.
The best-established application of molecular glues is inducing targeted protein degradation (TPD). By inducing PPI between a target protein and an E3 ligase, a ternary complex consisting of “target protein–molecular glue–E3 ligase” is created. This ternary complex formation induces the polyubiquitination of the target protein, leading to its degradation via the ubiquitin-proteasome pathway. This class of molecular glues are called molecular glue degraders. The immunomodulatory drugs (IMiDs), thalidomide and lenalidomide are two well-known examples of molecular glue degraders[1]. In recent years, both big pharma and biotech companies have expanded their efforts in developing molecular glues for targeted protein degradation (Figure 1).
Figure 1. Timeline of Major Molecular Glue Research and Development Milestones [1]
The major advantage of the TPD approach over traditional therapeutics lies in its ability to target a wider range of targets, especially “undruggable” ones. PROTACs (PROteolysis TArgeting Chimeras) and molecular glues are two major classes of TPD molecules. Compared to PROTACs, molecular glues have advantages in several aspects. For example, MGs have better physicochemical properties, lower affinity requirements, and reduced or no hook effect (Figure 2). These outperforming properties make MGs better candidates for drug development. However, the discovery of MGs is much more challenging than PROTACs, and the targets of MGs are difficult to manipulate.
Figure 2. PROTAC vs Molecular glues [2]
Molecular Glues Discovery
Molecular glue degrader discovery primarily relies on various screening strategies, often involving serendipitous discovery, high-throughput screening, or medicinal chemistry guided rational design[3,4,5]. The mechanism of action (MoA) for MGs involves inducing protein-protein interactions, which is often accompanied by conformational changes. As a result, the early discovery process of MGs typically has a low hit rate. This also complicates structure-activity relationship (SAR) analysis and structure-based drug design. Additionally, The MG hits are highly sensitive to chemical modifications. In light of these challenges, large-scale screening remains the mainstream approach for MG discovery.
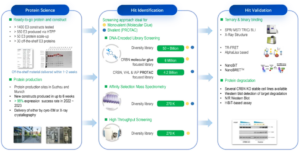
WuXi Biology has developed a comprehensive Molecular Glue Discovery Platform, equipped with DNA Encoded Library (DEL), Affinity Selection Mass Spectrometry (ASMS), and High Throughput Screening (HTS) technologies (Figure 3).
Figure 3. Molecular Glue Discovery Platform at WuXi Biology
DEL Screening
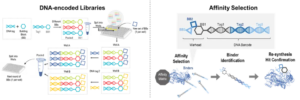
DEL is a collection of small molecule compounds generated through combinatorial chemistry, where each compound is attached to a unique DNA barcode to encode the identity of the compound. DNA-encoded libraries feature enormous compound diversity, low synthesis cost per molecule, and rich SAR information. The screening approach for DEL typically employs affinity-based screening, making them well-suited for molecular glue discovery (Figure 4).
Figure 4. DEL Screening Platform for Molecular Glue Discovery
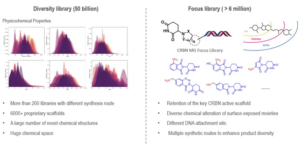
WuXi Biology offers two types of general, small molecule DELs for molecular glue discovery. One is a diversified library containing over 200 sub-libraries constructed by different synthetic routes, with over 50 billion structures. These compounds were designed based on over 6,000 bioactive scaffolds derived from public information, including drugs that are approved or in clinical phase, public databases, patents, or literature. These diverse libraries are ideal for targets with no reported molecular glues or ligands.
The other is molecular glue focused libraries, designed based on reported small molecule ligands or core scaffolds of molecular glues. This library significantly expands the chemical space of core scaffolds through extensive chemical modifications, while maintaining high target specificity and success rates. For example, WuXi Biology has developed an immunomodulatory imide drug (IMiD) focused library targeting the E3 ligase cereblon (CRBN). The solvent-exposed region of CRBN’s molecular glue is critical for substrate recognition and inducing PPIs. Extensive modifications were introduced to the solvent accessible interface by applying diverse synthetic routes and building blocks. This generated a focused library of approximately six million compounds in both solution and on-bead format. For targets with existing molecular glues and ligands, the focused library approach could secure a decent screening performance and hit rate (Figure 5).
Figure 5. Application of DEL for Molecular Glue Discovery at WuXi Biology
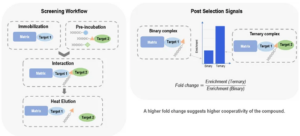
Different strategies are employed for molecular glue screening, depending on the format of the DEL library. For in-solution DEL, a pull-down based method with the presence of dual proteins is used for affinity selection. By comparing enrichment fold changes between single protein and dual protein conditions, potential molecular glue hits that induce ternary complex formation can be identified. A higher enrichment fold change indicates that the compound’s enrichment depends more on the presence of both proteins, suggesting a potential molecular glue interaction. (Figure 6).
Figure 6. Single Pull-down & Single Reaction for In-solution DEL
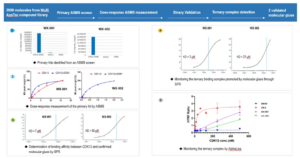
WuXi Biology utilized a known molecular glue compound, CC-885, which targets GSPT1/CRBN, as a reference compound for screening. CC-885 was conjugated to DNA as the molecules in the DNA-encoded libraries and spiked into the DEL library. The screening results showed that the DNA-conjugated CC-885 exhibited significantly higher enrichment when both GSPT1 and CRBN proteins were present, which validated the screening methods. More importantly, in the screen we observed several potential CRBN ligands that are highly enriched in the dual protein conditions. These molecules were further tested using HTRF assays, and the primary results suggested several potential MGs were identified (Figure 7).
Figure 7. In-Solution DEL Screening for GSPT1/CRBN Molecular Glues
For the on-bead DEL, the screening method also involves simultaneous incubation of both proteins. Two distinct fluorescently labeled antibodies (FITC and APC labelled) were used to detect the presence of the two proteins individually. The bead size is comparable to that of cells, thus compatible with flow cytometry. Fluorescence-activated cell sorting (FACS) is used to sort out the beads that show fluorescent signal shifts in both channels. The compounds on these beads are potential molecular glues that readily induce the ternary complex formation. The sorted beads can be sequenced by NGS to reveal compound structure. The CRBN-DDB1 molecular glue CC885 was used as a tool compound in a proof-of-concept screening. CC885 is mixed with negative on-beads compounds. FACS analysis robustly identified and sorted out CC885 signals, as exemplified by the populations with increased APC and FITC signals. Negative control compounds or blank beads, in contrast, remained unchanged (Figure 8). These results demonstrated the feasibility of using FACS as a robust semi-functional readout to identify potential molecular glue hits.
Similar screening against the IMiD focused on-beads library generated novel potent molecular glue molecules that have comparable potency with CC885 (data to be published soon).
Figure 8. On-Bead DEL Screening for CRBN/GSPT1 Molecular Glues
Affinity Selection Mass Spectrometry (ASMS)
ASMS is a widely used affinity-based screening method. Unlike DEL screening, the compound library for ASMS screening is shared with the conventional high-throughput-library, and they do not have DNA sequences attached to the compounds. Similar to the in-solution DEL screening method, molecular glue candidates in ASMS are identified by comparing recovered signals between single and dual proteins conditions.
ASMS molecular glue discovery at WuXi AppTec is empowered by an extensive small molecule library collection of over 270,000 compounds. In an attempt to search for novel MGs targeting the CDK12-CyclinK/DDB1 complex, we employed the ASMS strategy to screen a subset of 1000 compounds out of the full 270k libraries. Two compounds were identified as potential MG hits, which showed significant higher recovered signals in dual protein conditions. These compounds were validated in follow-up assays, including ASMS based dose-response titration, SPR, and HTRF (Figure 9).
Figure 9. Application of ASMS for Molecular Glue Discovery Targeting the CDK12-CyclinK/DDB1 Complex
HTS Screening
The one-compound-one-well high throughput screening (HTS) achieved by automation is a classical approach for hits discovery. With proper assays set-up, (e.g., Time-Resolved Fluorescence Resonance Energy Transfer, TR-FRET for detecting ternary complex formation, and degradation assays to evaluate degradation potency), HTS can be applied for novel molecular glue discovery. WuXi Biology has rich experience in applying HTS for molecular glue affinity screening and functional (or semi-functional) screening.
Comprehensive toolbox at WuXi Biology
With diverse screening capabilities as a leading drug discovery solution provider, WuXi Biology has developed a comprehensive toolbox to facilitate MG discovery, covering target identification, protein production, and compound validation using diverse biophysical and biochemical approaches, and crystallography (Figure 10). The integration of these highly related capabilities ensures a synergetic effort to expedite MG discovery
Figure 10. WuXi Biology Toolbox for Molecular Glue Discovery
References:
[1] Chem. Soc. Rev. 2022, 51, 5498–5517.
[2] Biochemistry. 2023, 62, 601−623.
[3] Science. 2017, 356, eaal3755.
[4] J. Med. Chem. 2021, 64, 1835−1843.
[5] J. Med. Chem. 2021, 64, 10606−10620.
WuXi AppTec | Targeted Protein Degradation Services:
- Learn about our extensive platform of services to support protein degradation research by clicking HERE
- View our flyer on Application of ASMS for Molecular Glue Discovery by clicking HERE
- View our White Paper, entitled: “Emerging Drug Discovery Strategies for Targeted Protein Degradation” by clicking HERE
- View our Poster entitled: “Biophysical & Functional Characterization of Bifunctional Small Molecules Enables TPD Drug Discovery” by clicking HERE
Related Content
With advancements in drug design, there is resurging interest in drugs that form covalent bonds with their targets. Covalent drugs...
VIEW RESOURCERNA-targeted therapeutics are highly intriguing due to their unique physiological properties and novel modes of action in drug R&D. Developing...
VIEW RESOURCE