Advances and Biological Evaluation of Oligonucleotide Drugs
Introduction:
As a therapeutic modality, oligonucleotides have a target abundance, persistent efficacy, high specificity, greater success rate and shorter R/D development period compared to other therapeutic approaches. Recently, significant achievements have been made in the discovery of oligonucleotide drugs. Numerous oligonucleotide drugs have been marketed. However, there are still many challenges, including limited delivery efficiency and off-target effects. Dr. Qiong Zhou, Executive Director of Discovery Biology of WuXi AppTec, shares her insights in the progress of oligonucleotides, as well as her team’s capabilities and experiences in the biological evaluations of oligonucleotides.
Outline of oligonucleotide drugs:
Oligonucleotides are chemically synthesized short (12 to 30 nucleotides) single or double-stranded DNA or RNA molecules. Oligonucleotides selectively regulate the cleavage, degradation and translation of target RNAs through Watson–Crick base pairing binding, consequently achieving the therapeutic purposes. Oligonucleotides include antisense oligonucleotides (ASO), small interfering RNAs (siRNA), aptamers and self-amplifying RNAs (saRNA). Currently marketed oligonucleotide drugs include ASOs and siRNAs. Table 1 and Figure 1 show the comparisons of ASO and siRNA.
Table 1. Comparisons of ASO and siRNA drugs
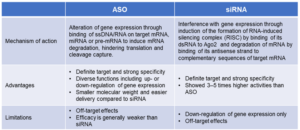
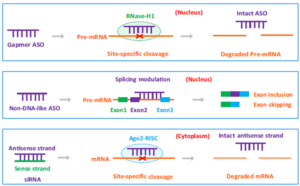
Figure 1. Comparisons of ASO and siRNA drugs [1]
Recent advances in the discovery of oligonucleotide drugs:
Nine ASOs and five siRNAs have been approved for the treatment of human diseases. Current efforts on the discovery of ASO and siRNA drugs mainly focus on improving the activity and reducing the toxicity (Table 2). Design of oligonucleotides is generally based on the sequence-specific binding to the target mRNAs. Significant experiences have been accumulated for the design of siRNAs. However, efficiency of ASO design is still relatively low and usually is required to synthesize more molecules to select a desirable molecule. Therefore, it needs to explore new technologies to improve design efficiency. However, the main challenges of the discovery of oligonucleotide drugs are limited delivery efficiency and potential toxicities. Oligonucleotides can be rapidly digested by nucleases and cleared by liver and kidneys, resulting in short half-lives and low concentrations in target tissues. In addition, oligonucleotides cannot freely traverse cell membrane due to their high molecular weights and negative charges. Furthermore, oligonucleotides can be trapped in endosomes, hence cannot play functions. The toxicity of oligonucleotides can be on-target and/or off-target effect. The off-target toxicity is mainly due to the nucleotide mismatches.
Table 2. Major challenges and strategies for the discovery of oligonucleotide drugs:
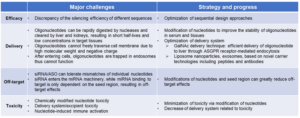
Remarkable efforts have been made to solve these challenges. By improvement of oligonucleotide designs and delivery methods, significant progresses have been achieved to reduce immunogenicity and to improve delivery efficiency of oligonucleotides.
Modifications of oligonucleotides need to consider to improve stability and to minimize immunogenicity, as well as to enhance pairing ability of bases (Figure 2). The first generation of ASOs was modified with a phosphorothioate (PS)-modified backbone to improve pharmacokinetic property and stability by reducing hydrophilicity and binding to plasma proteins and increasing resistance to nucleases [2]. However, phosphorothioate modifications can reduce the binding affinity of oligonucleotides to targets and can also lead to toxic effects after multiple administrations. The second generation of oligonucleotides was explored with various side chain and backbone modifications, such as replacement of 2ʹ-OH with 2ʹ-O-Me, 2ʹ-MOE or 2ʹ-F based on the PS-modified backbones. These modifications enhance stability and target binding affinity, and reduce immunogenicity as well. The third generation of oligonucleotides has modifications in sugar rings, including locked nucleic acid (LNA), peptide nucleic acid (PNA) and other changes which further improved stability and affinity of oligonucleotides.
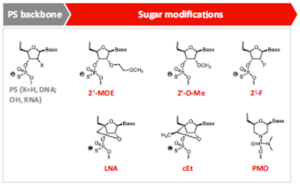
Figure 2. Structural modifications of oligonucleotides [2]
Chemical modifications are mainly directed at a single nucleotide. For large nucleic acids, it usually needs to take into account of combination of various and sequential modifications to achieve a satisfactory design. STC (standard template chemistry) and ESC (enhanced stabilization chemistry) modifications have been extensively explored (Figure 3). The principle of the STC is to modify 11 to 13 bp position at the 5′ end of an antisense chain with three consecutive methoxy moieties. While the principle of the ESC is to increase antisense chain to 23 nucleotides, to replace the first two phosphate bonds at the 5 ‘end of both sense and antisense chains with PS modifications, then to replace them with methoxy-modified ribose. Each oligonucleotide has a specific sequence, therefore it is not necessarily fully adapted to this modification template, but may need specific modifications.
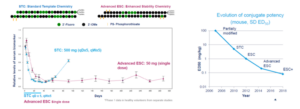
Figure 3. STC and ESC modifications [3]
Oligonucleotides are typically unable to reach their function sites by chemical modifications alone. In addition, an efficient delivery system is needed. Currently, the most common oligonucleotide delivery methods are based on lipid nanoparticle (LNP) and N-acetyl galactosamine (GalNAc) conjugation (Figure 4). LNP is a cationic lipid to improve transmembrane transport of oligonucleotides. Despite widespread clinical applications, LNPs still have some drawbacks, such as pro-inflammatory reactions. As a promising alternative approach, GalNAc delivery system exhibits high delivery efficiency and low potential toxicity [4]. GalNAc is a ligand targeting to asialoglycoprotein receptor (ASGPR), which is specifically and highly expressed on hepatocyte membrane. Hence, GalNAc has been successfully used for delivery of liver targeted oligonucleotides. However, it is highly needed to develop delivery methods for oligonucleotides efficiently targeting to non-hepatic tissues and cells.
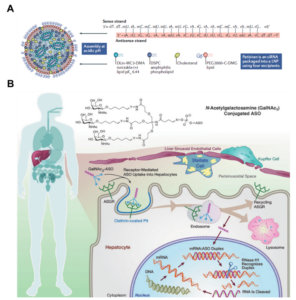
Figure 4. LNP (A) and GalNAc (B) delivery systems [5]
R&D process of oligonucleotide therapeutics:
Figure 5 summaries the general R&D approach of oligonucleotides. First, a suitable therapeutic target should be identified and validated. Second, a strategy for screening, nucleotide sequence design and modification as well as a delivery method should be established. Importantly, oligonucleotides are sequence-based drugs, therefore it is recommended to introduce the evaluation of off-target effects at an early stage. In addition, in vitro studies of free uptake and stability need to be carried out, followed with optimization of pharmacokinetic and toxicity profiles.
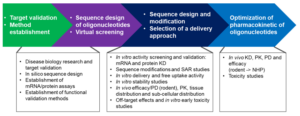
Figure 5. Research and development process of oligonucleotides
In terms of sequence design, it is essential to completely understand a target gene expression, isoform and SNP. Sequence differences across species also need to be considered for the correlation of results of animal studies to clinical outcomes. Generally, a designed sequence should at least represent human and monkey, ideally also represent the gene sequences of mouse, otherwise humanized mouse model may be needed for pharmacodynamic studies. In addition, it is necessary to analyze the overall off-target effects and to predict the activity using thermokinetics and sequence characteristics (Figure 6).

Figure 6. Considerations of design of oligonucleotide sequences
In vitro activity screening is to evaluate gene expression in cell-based assays by quantification of target mRNA and/or protein. However, overexpression of target gene with a stable cell line or a transient transfection approach may be needed for genes, especially for some disease-associated mutants which express at extremely low levels in normal cell lines, to ensure a successful screening [6].
The primary purpose of chemical modifications is to improve the activity of oligonucleotides. CADD (computer-aided drug discovery) can be employed for the chemical modifications (Figure 7).
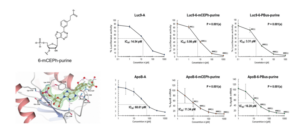
Figure 7. Effect of chemical modifications on the activity of oligonucleotides [7]
Another purpose of chemical modifications is to improve the stability and pharmacokinetic property of oligonucleotides. Oligonucleotide stability is commonly studied in in vitro assays in the presence of nuclease and serum. Furthermore, stability of oligonucleotides can be quantified with liver homogenate, which results generally have a better correlation across species and with gene knockdown activities in animal studies [8].
The third purpose of chemical modifications is to reduce immunotoxicity of oligonucleotides. Host immune responses can be induced by oligonucleotides via activation of pattern recognition receptor (PRR) of the innate immune. In vitro toxicity studies can be used to assess the effects of oligonucleotides on immune pathways and downstream cytokine release and immune cell activation. In addition, oligonucleotides have potentiality to activate the complement system. It is recommended that species differences should be considered for the evaluation of complement activation.
The binding ability of a delivery system and receptor can be evaluated by surface plasmon resonance (SPR) or cell-based assay approaches (Figure 8). Determination of EC50 values can be carried out with fluorescence labeling to quickly and conveniently evaluate the effectiveness of a delivery system.
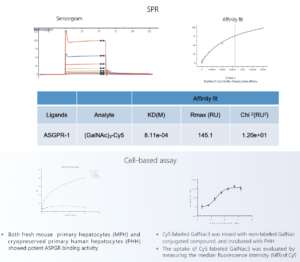
Figure 8. In vitro evaluation of delivery efficiency
Efficiency of a delivery system can be assessed by free uptake assays [9]. Oligonucleotides can enter cells with a self-delivery system without a chemical transfection reagent, subsequently delivery efficiency and oligonucleotide activity can be evaluated by quantifying a target mRNA and/or protein (Figure 9).
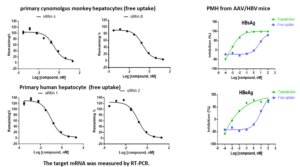
Figure 9. Assessments of delivery efficiency of oligonucleotides in primary hepatocytes by free uptake assays
Tissue distribution and pharmacokinetic property of an oligonucleotide should also be studied. Concentration of an siRNA in RISC (RNA-induced silencing complex) was observed to have close correlation with its pharmacodynamic effect. Therefore, it is essential to maintain the binding ability of a modified oligonucleotide to RISC proteins, such as Ago2 (Argonaute RISC catalytic component 2). In vitro and/or in vivo studies should be considered to assess the RISC bind and efficacy of oligonucleotides (Figure 10).
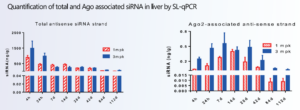
Figure 10. Quantifications of the total and RISC Ago-associated siRNAs [10]
Although sequence specificity is considered in the sequence design process, it is difficult to guarantee that the designed sequence can be directly tested with normal mice because of the sequence differences across species. Therefore, “humanized” mouse models are needed to study the in vivo efficacy of oligonucleotides. Hydrodynamic injection (HDI) mouse models are frequently used for evaluation of liver-target oligonucleotides. Adeno-associated virus (AAV) animal models can be used for study of oligonucleotides targeting liver or extrahepatic tissues. Establishment of an AAV animal model is relatively fast, but packaging size of an AAV vector is limited. Transgenic animal models can also be used for evaluation of liver and extrahepatic targeted oligonucleotides for both pharmacodynamics and disease related readouts, but generation of a transgenic animal model is time consuming and expensive. In addition, humanized mouse models can be used for evaluations of both on-target and off-target effects of oligonucleotides. Generally, the in vivo efficacy of oligonucleotides is initially studied in mouse models, then confirmed in monkey models.
Different strategies should be taken at different stages for study of off-target effects of oligonucleotides (Figure 11). At early stage, in-silico tools can be used for preliminary sequence analysis. Upon identification of lead oligonucleotides with desired activities in in vivo studies, RNA-seq technology should be employed to identify a potential off-target gene(s). In addition, psiCheck reporter system is a fast and effective tool to evaluate off-target effects by simulating off-target sequences in the early modification/screening stage.

Figure 11. Strategies for the evaluation of off-target effects of oligonucleotides
Prospects of oligonucleotide drugs:
Oligonucleotides have numerous potential advantages compared with small molecules and antibodies. Key advantages include abundant targets, high targeting specificity and long-acting effect. Digital drug design tools for oligonucleotides are readily available. The rapid and relatively straight-forward chemical synthesis process and production of oligonucleotides confer obvious advantages over antibodies. Furthermore, with the continuous improvement of delivery methods and chemical modifications, the clinical applications of oligonucleotides are expected to become more and more prevalence.
Future R&D directions of oligonucleotide drugs include identification of new liver targets and development of more efficient delivery technologies for extrahepatic organs, as well as elucidation of endosome escape mechanisms and minimization of toxic effects [12]. Furthermore, breakthrough of chemical modification bottlenecks could lead to the discovery of the next generation of oligonucleotide therapeutics with enhanced efficacy and reduced toxicity.
WuXi AppTec oligonucleotide service platform:
The Discovery Biology Unit of WuXi AppTec has a comprehensive platform for support of the discovery of oligonucleotide drugs. Our services include in vitro screening, in vivo activity assessment, design of delivery system and evaluation of early toxicity. By integration of our biology and chemistry capabilities, we can offer end-to-end solutions to support our collaborators for acceleration of their oligonucleotide drug programs (Figure 12).
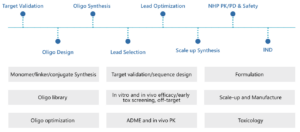
Figure 12. WuXi AppTec oligonucleotide capabilities
References:
- Yin W, Rogge M. Targeting RNA: a transformative therapeutic strategy. Clin Transl Sci. 2019 Mar;12(2):98-112. doi: 10.1111/cts.12624.
- Crooke ST, Baker BF, Crooke RM, Liang XH. Antisense technology: an overview and prospectus. Nat Rev Drug Discov. 2021 Jun;20(6):427-453. doi: 10.1038/s41573-021-00162-z.
- Information disclosed by Alnylam. https://www.alnylam.com/.
- Paunovska K, Loughrey D, Dahlman JE. Drug delivery systems for RNA therapeutics. Nat Rev Genet. 2022 May;23(5):265-280. doi: 10.1038/s41576-021-00439-4.
- Crooke ST, Baker BF, Xia S, Yu RZ, Viney NJ, Wang Y, Tsimikas S, Geary RS. Integrated assessment of the clinical performance of GalNAc3-conjugated 2′-O-methoxyethyl chimeric antisense oligonucleotides: I. Human Volunteer Experience. Nucleic Acid Ther. 2019 Feb;29(1):16-32. doi: 10.1089/nat.2018.0753.
- Wang Q, Yin H, Camelliti P, Betts C, Moulton H, Lee H, Saleh AF, Gait MJ, Wood MJ. In vitro evaluation of novel antisense oligonucleotides is predictive of in vivo exon skipping activity for duchenne muscular dystrophy. J Gene Med. 2010 Apr;12(4):354-64. doi: 10.1002/jgm.1446.
- Schmajuk G, Sierakowska H, Kole R. Antisense oligonucleotides with different backbones. Modification of splicing pathways and efficacy of uptake. J Biol Chem. 1999 Jul 30;274(31):21783-9. doi: 10.1074/jbc.274.31.21783.
- Basiri B, Xie F, Wu B, Humphreys SC, Lade JM, Thayer MB, Yamaguchi P, Florio M, Rock BM. Introducing an in vitro liver stability assay capable of predicting the in vivo pharmacodynamic efficacy of siRNAs for IVIVC. Mol Ther Nucleic Acids. 2020 Sep 4;21:725-736. doi: 10.1016/j.omtn.2020.07.012.
- Shinohara F, Oashi T, Harumoto T, Nishikawa T, Takayama Y, Miyagi H, Takahashi Y, Nakajima T, Sawada T, Koda Y, Makino A, Sato A, Hamaguchi K, Suzuki M, Yamamoto J, Tomari Y, Saito JI. siRNA potency enhancement via chemical modifications of nucleotide bases at the 5′-end of the siRNA guide strand. RNA. 2021 Feb;27(2):163-173. doi: 10.1261/rna.073783.119.
- Zhang R, Calixto CPG, Marquez Y, Venhuizen P, Tzioutziou NA, Guo W, Spensley M, Entizne JC, Lewandowska D, Ten Have S, Frei Dit Frey N, Hirt H, James AB, Nimmo HG, Barta A, Kalyna M, Brown JWS. A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucleic Acids Res. 2017 May 19;45(9):5061-5073. doi: 10.1093/nar/gkx267.
- Kobayashi Y, Fukuhara D, Akase D, Aida M, Ui-Tei K. siRNA seed region is divided into two functionally different domains in RNA interference in response to 2′-OMe modifications. ACS Omega. 2022 Jan 4;7(2):2398-2410. doi: 10.1021/acsomega.1c06455.
- Roberts TC, Langer R, Wood MJA. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov. 2020 Oct;19(10):673-694. doi: 10.1038/s41573-020-0075-7. Epub 2020 Aug 11.
WuXi AppTec | Oligonucleotide Services:
- Learn about our extensive panel of oligonucleotide drug discovery services by clicking HERE
- Read our White Paper entitled: “Oligonucleotide Therapeutics: Drug Discovery Through Tightly Coupled Chemistry and Biology” by clicking HERE
- Learn about our platform: Anti-Tumor Nucleic Acid Drug Discovery by clicking HERE
Related Content
From April 25 to 30, 2025, the annual meeting of the American Association for Cancer Research (AACR), one of the...
VIEW RESOURCEOncoWuXi Express will continue to keep you informed about updates to our online tumor model database (OncoWuXi Database), as well...
VIEW RESOURCE
