mRNA Display: Unlocking Peptide Discovery
Introduction:
In the realm of modern therapeutics, peptides have emerged as a powerful and versatile class of molecules, unlocking their potential through continuous advancements in innovative technologies. Among these breakthroughs is mRNA display, a technique that has accelerated peptide discovery. This article explores the principles and key steps of mRNA display, as well as its application in peptide discovery and screening.
The Rise of Peptide Therapeutics
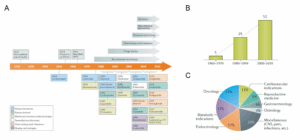
Since the early 20th century, peptides have played a pivotal role in disease treatment. The discovery of insulin in the 1920s revolutionized the treatment of type 1 diabetes, marking a significant milestone. This was followed by the introduction of cyclosporine in the 1980s. As of 2019, over 80 peptide drugs had been approved for various medical applications. By the end of 2024, this number had approached 100 (Figure 1). This rapid growth underscores the increasing importance and potential of peptide-based therapies [1, 2].
Figure 1. Historical timeline of key milestones, developments, and drug approvals in the peptide therapeutics field [1].
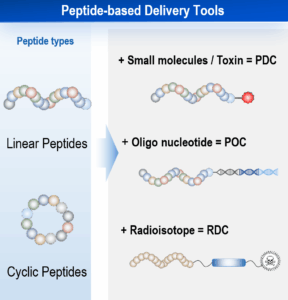
Peptide drugs possess a unique set of characteristics that place them between small molecules and large biologics. Unlike small molecules, peptides can target previously “undruggable” binding sites, offering new therapeutic possibilities. They demonstrate high specificity, low immunogenicity, and are relatively easy to synthesize and modify. However, peptides also face challenges, such as poor permeability and limited oral bioavailability [3-6]. Despite these hurdles, advancements in medicinal chemistry and innovative technologies have paved the way for the development of cell-permeable and orally available peptides (Figure 2A). Beyond their direct therapeutic use, peptides offer significant potential to synergize with other therapeutic modalities through diverse conjugation strategies. Key examples include Peptide-Drug Conjugates (PDCs), which link peptides to cytotoxic payloads; Peptide-Oligonucleotide Conjugates (POCs), combining peptides with nucleic acid therapeutics; and Peptide Radioligand Conjugates (RDCs), also known as Peptide-Radionuclide Conjugates (PRCs). RDCs specifically leverage a targeting peptide linked to a diagnostic or therapeutic radioisotope. This design enables highly precise delivery of radiation to cells expressing the target receptor (Figure 2B).
Figure 2. The characteristics of peptide drugs and peptide-based delivery tools [3-6].
The Principles and Advantages of mRNA Display
mRNA display is a powerful technique that leverages the natural process of translation to generate and screen large libraries of peptides. This method involves the in vitro translation of an mRNA library, during which peptides are synthesized and covalently linked to their encoding mRNA through a puromycin tag. The resulting peptide-mRNA complexes can then be subjected to various selection processes to identify peptides with desired binding properties [7].
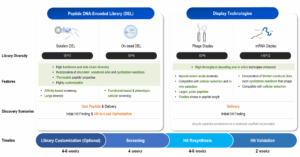
At WuXi Biology, we offer a range of peptide discovery technologies, including peptide DNA-Encoded Library (DEL), phage display, and mRNA display. Our peptide DEL library possesses a diversity of around 10^11, and with peptide technology, we can incorporate a large number of unnatural amino acids. If a larger macrocycle is desired, phage display and mRNA display are superior options. However, phage display has several disadvantages compared to mRNA display. For one, phage display has lower diversity and is performed in vivo rather than in vitro. Expressing peptides on the phage surface comes with numerous limitations. Compared to traditional methods, such as phage display, mRNA display offers several advantages. It allows for the creation of libraries with greater diversity, often exceeding 10^12 unique sequences. This vast diversity increases the likelihood of identifying high-affinity binders. Additionally, mRNA display is performed in vitro, eliminating the limitations associated with in vivo expression inherent to phage display. This flexibility also allows for the incorporation of a wide range of unnatural amino acids, although natural amino acids are often sufficient for initial screening (Figure 3).
Figure 3. Comparison of DEL and display technologies.
mRNA Display Workflow
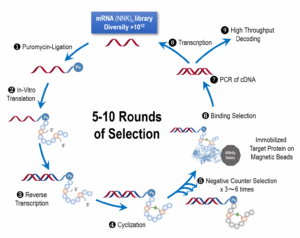
The workflow of mRNA display begins with the creation of an mRNA library with randomized sequences. These sequences are designed to cover peptides of varying lengths, typically ranging from 6 to 15 amino acids. The mRNA is then ligated to puromycin, which stalls translation upon entry into the ribosome. During in vitro translation, the ribosome reads the mRNA sequence, synthesizes the peptide, and attaches the puromycin to the peptide’s terminus. This process links the encoding mRNA to the peptide, facilitating downstream amplification and selection.
The next step involves cyclization of the peptide, which can be achieved through various linkers or cysteine-cysteine disulfide bonds. The cyclized peptides are then subjected to selection against an immobilized protein target. This process often includes counter-selection to remove non-specific binders, followed by multiple rounds of selection to enrich for high-affinity binders. Finally, the enriched sequences are analyzed through high-throughput decoding to identify promising candidates (Figure 4).
Figure 4. mRNA display workflow.
Case Study
Below, we present a case study of mRNA display screening at WuXi Biology. We employed a multi-round selection strategy to screen and identify biotin-target binders. Figure 5 illustrates the selection strategy and performance assessment results.
In the first round of selection, the peptide library was incubated with the target to screen for initial binders. Subsequent rounds 2-6 involved negative selection to eliminate non-specific binders, followed by positive selection to enrich for specific binders. In the seventh round, we further distinguished orthosteric and allosteric binders. By introducing a known ligand after negative selection, we were able to detect allosteric binders. Conversely, introducing the protein target without the ligand allowed us to identify orthosteric binders.
The experiment results show the recovery trend in selection performance, indicating that the enrichment of the target significantly improved with increasing selection rounds. Additionally, the identity matrix with a score of > 40% suggests that the selected peptides have high specificity. The heatmap and sequence logo further reveal 19 sequence clusters, with more than 10 peptides in each, demonstrating good clustering effects. Ultimately, 21 peptides with good performance were selected from these clusters for validation. These results indicate that our method effectively screens and identifies binders with strong specificity.
Figure 5. Case Study of mRNA Display Screening Performance.
Future Directions
As peptide therapeutics continue to gain prominence, mRNA display and related technologies will play a crucial role in driving innovation. The ability to rapidly screen large libraries of peptides and identify high-affinity binders unlocks new possibilities for drug development. With advancements in library design, selection techniques, and data analysis, the potential for discovering novel peptide therapeutics is limitless.
In conclusion, mRNA display has emerged as a powerful tool in peptide discovery, offering a unique blend of high diversity, flexibility, and efficiency. Its integration with peptide DNA-encoded libraries and other advanced platforms further enhances its potential. As researchers continue to explore the vast landscape of peptide therapeutics, mRNA display will undoubtedly remain at the forefront of this exciting field.
WuXi Biology Integrated Peptide Discovery Platform
WuXi Biology provides multiple approaches to discover peptide hits, including DEL and display technologies. Peptide DELs are synthesized through chemical reactions, whereas display technologies utilize biological synthesis. The combination of DEL and display technologies allows clients to explore a vast chemical space, unlocking new possibilities for peptide discovery.
References
- Muttenthaler, Markus et al. “Trends in peptide drug discovery.” Nature reviews. Drug discovery vol. 20,4 (2021): 309-325.
- https://www.fda.gov/
- De Groot, Anne S et al. “Immunogenicity risk assessment of synthetic peptide drugs and their impurities.” Drug discovery today vol. 28,10: 103714 (2023).
- Chang, Liwei et al. “Towards rational computational peptide design.” Frontiers in bioinformatics vol. 2 1046493. 21 Oct. (2022)
- Pei, Jingyan et al. “Advances in the stability challenges of bioactive peptides and improvement strategies.” Current research in food science vol. 5 2162-2170. 5 Nov (2022).
- Gómez-Guerrero, Néstor Alejandro et al. “Synthetic Peptides in Doping Control: A Powerful Tool for an Analytical Challenge.” ACS omega vol. 7,43 38193-38206. 21 Oct (2022).
- Josephson, Kristopher et al. “mRNA display: from basic principles to macrocycle drug discovery.” Drug discovery today vol. 19,4: 388-99 (2014).
Related Content
In this webinar, WuXi AppTec scientists discuss the latest innovative technologies and products in early-stage drug discovery. Viewers will gain...
VIEW RESOURCEPeptide-based therapeutics have emerged as a significant class of drugs in the pharmaceutical landscape, offering numerous advantages, including high specificity,...
VIEW RESOURCE