Enabling Technology for Targeted Protein Degradation

Summary
Traditional small molecule therapeutic mechanisms have recently been augmented by a new strategy to specifically manipulate the levels of disease-related proteins. By employing bifunctional molecules, we can hijack endogenous cellular degradation mechanism for targeted degradation of a disease-related protein. Bifunctional molecules consisting of a ligand that binds to an E3 ligase, connected by a linker to another ligand that binds to the disease-related protein, are often referred to as degraders. This approach may render some previously “undruggable” targets amenable to therapeutic intervention.
During this webinar, one academic and two subject matter experts from WuXi AppTec illustrated the past, current state-of-the-art and the future of these exiting chemical modalities in drug discovery. Lessons learned, opportunities and challenges have been debated in detail. Depicted by several examples they demonstrated the power of integrated drug discovery platform, which encompasses high quality protein production, direct binding screens like fragment and DEL screens, biophysics, structural biology and in vitro pharmacology.
Key Learning Objectives
- Identification and characterization of new bifunctional molecules including degraders using synergistic power of fragment and DEL screening, biophysics and structural biology
- Chemical and computational design strategies for degraders
- In vitro pharmacology tools
- Strategies and tools provided by WuXi AppTec protein degrader platform
Speakers

- William Farnaby, PhD
- Team Leader, Chemical Biology and Medicinal Chemistry, University of Dundee
- Will Farnaby is currently a team leader in medicinal chemistry and chemical biology at the School of Life Sciences, University of Dundee. Since 2016 he has led a multi-disciplinary team of drug discovery scientists engaged in a collaboration between Professor Alessio Ciulli and Boehringer Ingelheim. This unique collaboration model aims to discover bi-valent degraders to address areas of critical unmet medical need. William has led and progressed protein degradation projects through multiple stages of the drug discovery process and in doing so continues to contribute significantly to the advance of rational degrader design, synthesis and methods of biochemical, biophysical and cellular evaluation. Prior to this William worked for 8 years at Takeda Cambridge as a senior medicinal chemist where he contributed to the discovery of multiple pre-clinical and clinical candidates including CH24H inhibitor Soticlestat.

- Dave Madge
- Vice President, Research Service Division, WuXi AppTec
- Dr Dave Madge is Vice President of the Research Services Division in WuXi AppTec. In this role Dave leads a portfolio of drug discovery initiatives across multiple therapeutic areas and modalities. Prior to joining WuXi AppTec, in 2014, Dave was VP, Research, for the ion channel drug discovery company Xention Ltd, in Cambridge, UK, developing new molecules for cardiovascular and respiratory disorders. Before Xention Dave was based at University College London, as part of an integrated biomedical research group, and was responsible for developing new therapeutic discovery projects into funded biotech companies. Dave has a PhD in Medicinal Chemistry from Imperial College, London, and started his career at The Wellcome Foundation in the medicinal chemistry team.

- Nuska Tschamer
- Head of DEL Lab Operations, Crelux, a WuXi AppTec Company
- Dr. habil. Nuska Tschammer is Head of DEL Lab Operations at WuXi AppTec´s HitS/Crelux in Munich, Germany. Prior to this role, she was Subject Matter Expert at Crelux. Before that she was Head of Research and Development Biochemistry at NanoTemper Technologies, where she developed new generation of dyes for the MicroScale Thermophoresis (MST) and Dianthus (TRIC). During the habilitation in Medicinal Chemistry at Friedrich-Alexander University, Erlangen, Germany, her research on the allosteric modulation of GPRCs was funded by several grants of German Research Foundation (DFG), which led to many publications and patents. She received various prizes and awards, among them Innovation Award in Medicinal Chemistry, which is jointly awarded by Society of German Chemists (GDCh) and German Pharmaceutical Society (DPhG).
Content:
PART 1
Birth of a new modality– Bifunctional molecule
Bifunctional molecule is molecule, which specifically binds two or more proteins and chemically induces the proximity of target proteins, which results in new biological effects (e.g. protein degradation, complex stabilization).
As a fascinating modality, nature got there before people. Darwin first postulated the existence of molecule that was released in plants which stimulated growth direction. This molecule called “Auxin” was isolated by plant physiologist Kenneth V. Thimann in 1942. Perhaps auxin was the first molecular glue to be characterized. However, people had to wait until 2007 before the mechanism of action (MoA) was fully understood that auxin could regulated the binding of TIR1-Aux/IAA. Auxin binds the TIR1 subunit of ubiquitin ligase SCFTIR1, and enhances docking of substrate proteins. Normally these proteins dissociate from TIR1 before they can be modified. However, TIR1 and auxin form a complementary surface that stabilizes the association of substrates, which undergo ubiquitylation followed by proteasomal degradation.
The example of natural glues helps scientists understand the MoA for a class of drugs, like thalidomide. Thalidomide was invented in the 1950s and once widely used as a drug to relieve morning sickness in pregnant women, but was withdrawn from the market after it was discovered to cause severe birth defects. 40 years later the reason for teratogenicity of thalidomide was identified. This drug binds to CRBN and changes its recognition of the substrates that can be degraded. This results in the degradation of transcription factor Sall4, the accumulation of MEIS2 and the destabilization of CD147/MCT1, which leads to fetal malformations and organ defects. This example illustrates the importance of a deep understanding of the selectivity of natural glues and their impact on the cell signaling machinery.
Nowadays, scientists are able to construct bifunctional molecules through interwoven state-of-the art methods of chemistry, biophysics and structural biology. In general, bifunctional molecule is composed of warhead, linker and selective e.g. ligase binder to bring into close proximity two proteins that normally would not interact. This chemically induced proximity may result in specific control of biological processes like transcription, signaling cascades, protein folding and degradation. Targeting “undruggable” proteins provides us with new ways to investigate functions of specific protein and create new therapeutic strategies.
What is PROTAC® ?

PROTAC (PROteolysis TArgeting Chimera) is a bifunctional molecule composed of an E3 ligand that binds to an E3 ligase, a ligand that binds to the protein of interest, and a linker connecting the two ligands. A group of proteins, called E3 ligases, promote protein ubiquitination and degradation by tag them with ubiquitin protein molecules. As a part of natural protein disposal mechanism, the ubiquitinated protein is designated for degradation by the ubiquitin proteasome system. The PROTAC molecule is released and can continue its degradation mission.
Key events in the timeline
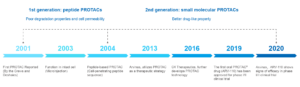
PROTAC approach aroused attention already nearly 20 years ago. Although the performance of the first generation of peptide degraders was rather limited, it was fully sufficient as a proof-of-concept that opened a great avenue of opportunities. Over the next years the scope of this technology continued to expand and to overcome many limitations. The second generation of degraders was chemically improved and had better drug-like properties. For example, the pipeline of Arvinas exploits targeted protein degradation as a therapeutic strategy since 2013. In 2019, the first oral PROTAC® drug (ARV-110, targeting androgen receptor for degradation) has been approved by FDA for phase I/II clinical trial in treating patients with metastatic castration resistant prostate cancer. In May 2020 Arvinas announced that ARV-110 to show signs of efficacy in these patients, and thus providing a strong validation of their PROTAC® technology.
Opportunities and challenges
Protein degraders contain two small molecule-binding ligands joined together by a linker. This new modality moves away from so called ‘rule of five’ in chemistry and redefines the definition of small molecule drugs. Preclinical and clinical studies demonstrate that protein degraders can be introduced into body by multiple routes of administration including oral. Besides, they can also be widely distributed to organs and tissues including brain.
In any new areas, opportunities always come with challenges. Discovery of protein degraders remains to be challenging due to the following factors and open questions:
- Discovery of protein degraders remains empirical and little rationale guiding the pairing of a specific E3 ligase with a given protein target
- What percentage of a target protein should be degraded to trigger a phenotypic response?
- Is acquired resistance to PROTAC treatment a possibility?
- Will PROTAC degrade off-targets that were untouched by their parent inhibitors?
With this in mind, the technologies need to be developed, which will facilitate better understanding of how these bifunctional molecules behave and how they work.
PART 2
Key tools and strategies available at WuXi AppTec
The identification and development of bifunctional compounds can be facilitated by the use of novel screening methods, state-of-the art biophysical tools and structural biology. The selection of most effective screening method and supporting methods depends on various factors.
Identification and development of bifunctional compounds
In contrast to classical drug pharmacology, functional activity is not necessary for either ligand in the bifunctional protein degrader. The moderate to high affinity ligands targeting the protein of interest and/or the E3 ligases are perhaps best found by direct binding screens, such as fragment based or DNA-encoded library (DEL) screens. In order to achieve full potential of protein degraders in drug discovery, WuXi AppTec has a rich toolbox enabling hit finding and hit-to-lead optimization including but not limited to DNA-encoded library (DEL) and fragment screening, structure-based and computationally-assisted design of degraders.
Identification of bifunctional compounds by DEL screening
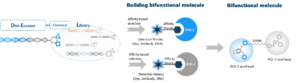
In WuXi AppTec DNA Encoded Library, about 90 billion chemical compounds are produced from over 100 selective hot cores for latent druggability, about 6 000 WuXi AppTec proprietary scaffolds and about 35 000 commercially source building blocks. All compounds are tethered to the DNA barcode. At the end of the affinity selection process, the compounds which remain bound to the protein of interest can be easily identified by decoding the unique barcode. Ligands discovered by DEL screening always possess a solvent accessible vector (this vector is the residual linkage connecting the small-molecule to its DNA barcode), and this vector may serve as a chemical handle for the addition of a bifunctional degrader linker.
Overall, DEL screening has the following advantages:
- Affinity-based selection that ensures physical binding
- Unprecedented library scale and diversity allows for high coverage of novel chemical space
- Already-in-place linker eliminates hassles in linker design
- Minimal protein consumption (5 µg/condition), rapid process (4 weeks) and parallel conditions for quick survey of multiple targets/conditions
Beyond that, WuXi AppTec DEL library can be customized and built according to different requirements and needs. Importantly, at the end of the DEL screening campaign, WuXi AppTec offer the synthesis without the DNA-tag (so called off-DNA compounds) and full hit confirmation by biophysical and in vitro assays.
Identification of unique binding pockets by fragment screening
Fragments here are small molecules with molecular weight usually less than 250 Da. Because of their small size, they have an ability to identify unique binding pocket on the protein surface. A diverse set of 1 000 fragments represents its chemical space about as effectively as would 10 trillion diverse drug-sized molecules. Also, once with a fragment in hand, scientists have ability to optimize pharmacokinetic profile simultaneously with potency as fragment hit grows. Generally, this development is enabled by the structural information obtained through the co-crystallization of fragments and the target protein. Still, about 30 % of all fragment screening campaigns continues to the next level without any structure information, and simply followed by old-fashioned chemistry campaign.
WuXi AppTec fragment library contains about 3 000 fragments, which were carefully selected to fulfill the strict requirements. For the detection of fragment binding, it is recommended to use state-of-the-art biophysical methods, because only the biophysical methods are sensitive enough to detect the binding between the fragment and the protein of interest.
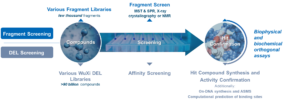
In conclusion, synergistic use of DEL screening and fragment screening enables discovery of novel ligands and bifunctional compounds. State-of-the art biophysical platform at WuXi AppTec HitS enables detailed characterization of their binding properties and MoA.
Structural elucidation by X-ray crystallography
High-resolution ternary complex crystal structures and biophysical investigations can guide rational and efficient optimization bifunctional protein degraders. Modern structure-based drug design (SBDD) relies not only on structural information but also on information on the thermodynamics, kinetic, and dynamics of target-ligand interactions. Because of unprecedented resolution and throughput, structural elucidation by X-ray crystallography is still the main choice.
Structure-based design of bifunctional molecules
Structure-based design of bifunctional molecules can yield chemically very different ligand as exemplified by the design of MacroPROTAC-1. In this case the crystal structure of MZ1 in complex with Brd4BD2 and VHL was obtained at first. As the vicinity of the binding mode of Brd4 ligands is clear, there was idea that one could actually use macrocyclization strategy to practically change this ligand. This macrocyclization strategy turned out to be very successful and the group obtained potent degrader named MacroPROTAC-1. Despite it lost some affinity at Brd4, it still yields a potent, fast and selective Brd4 degrader.
In Silico modeling of PROTAC-mediated ternary complexes conformation
There are 3 methods for PROTAC conformation mimicked in Silico modeling:
Method 1: PROTAC conformations are sampled independently, followed by post hoc addition of rigid body proteins
Method 2: The PROTAC is sampled in the context of one of the proteins, with the second added afterward
Method 3: PROTAC conformations are sampled independently of the proteins (and, indeed, a conformational database from Method 2 can be used, or vice versa), but possible ligase-target arrangements are provided via protein−protein docking
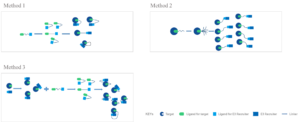
While further crystal structure assays will surely follow, scientists have demonstrated that the methods in this work, particularly the protein−protein docking-based Method 3 are accurate enough to complement ongoing structural campaigns. All of the methods offer reasonable and crystal-like geometries for ternary complexes, which can serve as starting point for testing hypotheses and refining principles of PROTAC design.
Workflow of in Silico PROTAC Platform
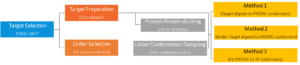
WuXi AppTec has developed computational workflows for preparation of target structure which is fit for biomolecules or to other proteins. As well as for linker selection and sampling of the many available conformers. Considering target interactions with bifunctional molecule, candidate ligase interaction with bifunctional molecules as well as ternary complexes, this platform can bring these data sets together with three main conformation methods.
WuXi AppTec Chemical Services Unit (CSU) platform also offers a comprehensive chemistry toolbox, in which frequently used E3 ligase ligands are readily available, like VHL, CRBN, MDM2, IAP. More than 50 most frequently used linkers are prepared, including PEG/Alkyl/”click-reaction” chain/rigid linkers. And more than 100 precursors (linker+E3 ligand) based on literature are readily to use. Recently, multi-kilo and GMP synthesis and formulation of these molecules has been carried out.
Characterization of Binding Properties
Once the bifunctional molecules are at hand, detailed investigation of binding properties is required. Crucial aspect of this analysis is the characterization of the ternary complex formation. How to choose appropriate biophysical and biochemical methods for ternary complex formation assay?
Biophysical characterization of binding properties by SPR
Protein degraders offer potential for improved selectivity beyond that of the constituent target ligand by harnessing additional stabilizing or destabilizing de novo protein–protein or protein–linker interactions formed via the ternary complex. Furthermore, the binding of the bifunctional molecule to the one protein partner (POI-I) may be enhanced or reduced by the presence of the secondary binding partner (POI-2). This can occur in the opposite order, where bifunctional molecules first interact with POI-2 to form a binary complex, and then it will interact with POI-1.
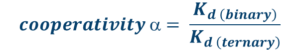
This effect can be quantified in terms of a “cooperativity” (a) factor. Biophysical methods recommended for the characterization of binding properties are Surface Plasmon Resonance (SPR; kinetics, binding affinity), MicroScale Thermophoresis (MST; binding affinity), Temperature-Related intensity Change (TRIC/Dianthus; binding), and X-ray crystallography (structural information).
Biophysical characterization of binding properties by MST/TRIC
MicroScale Thermophoresis (MST) is a powerful technique to quantify biomolecular interactions.
In this technique a variation in the fluorescence signal is detected, which is a result of a temperature gradient induced by an infrared laser. The extent of the variation in the fluorescence signal correlates with the binding of a ligand to the fluorescent target.
For example, during the formation of CRBN1/DDB1/GSPT1 complex, PROTAC CC-885 showed strong positive cooperativity and thus significantly higher affinity for the ternary complex as seen from the shift of the dose-response curve.
Characterization of protein degraders using in vitro assays
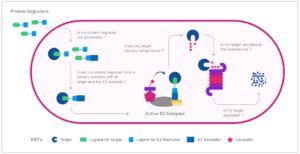
The picture depicts all the key stages in the cellular protein degradation mediated by protein degraders. We have to know if the protein degrader is able to penetrate the cell membrane and enter the cell. Then, once in the cell, we need to know whether a ternary complex composed of protein degrader, the target and the E3 recruiter is formed. In the next step we have to determine the activity of the ternary complex and if the target protein becomes ubiquitinated. Finally, we have to determine, is our target protein gets degraded by proteasome. And HD Biosciences (HDB) from WuXi AppTec has assays established, which cover all these steps of targeted protein degradation.
NanoBRET for E3-PROTACTM Binary Binding Assay
The E3-PROTAC cellular engagement assay (NanoBRET) is a powerful tool to evaluate the cell permeability and the engagement kinetics of protein degrader. In this live-cell assay, NanoLuc® luciferase is fused to VHL as the energy donor and a fluorescent cell-permeable small molecule (VHL tracer) serves as the energy acceptor. Protein degraders that are cell-permeable and engage VHL will compete with the tracer for binding to VHL, causing a decrease in BRET signal.
AlphaLISA for Target-PROTAC-E3 Ternary Complex Formation Assay
AlphaScreen/AlphaLISA assay is recommended for biochemical ternary complex formation assay due to relatively large complex formation involved. For ternary complex formation assay, AlphaLISA is mainly employed as a sandwich immunoassay. A biotinylated anti-VHL E3 ligase antibody binds the donor bead while a second anti-Brd4 bromodomain antibody is conjugated to AlphaLISA acceptor beads. In the presence of the large complex, beads come into close proximity. Donor bead excitation releases singlet oxygen molecules that transfer energy to the acceptor beads with light emission at 615 nm or 680 nm. Key of ternary biochemical assay development includes selection of potent reference compound, usage of high-quality protein and identifying proper detection condition.
HiBiT for Target Degradation Assay
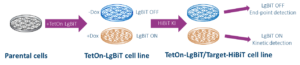
This HiBiT Platform encompasses a ‘2 in 1’ system, which enables monitoring of target protein degradation at physiological level and supports HTS/SAR screening and protein degraders kinetics study on the same cell line. Choosing an end-point assay is the most applicable method when we want to analyze a larger set of compounds in a short period of time. Based on these data we can than choose the most interesting compounds and go to the next step. In this step we can perform kinetics assay to obtain all parameters of interest. Because both type of assays are performed in the same cell line, the comparison of the data is easy.
PART 3
What are the future development trends for bifunctional molecules?
Above described assays (either biophysical or biochemical) were originally employed to characterize bifunctional protein degraders like PROTAC. Nowadays, these assays are applied to characterize the activity of diverse bifunctional molecules, which go beyond classical ubiquitin-mediated protein degradation. Here are some ideas and suggestions.
Extending the repertoire of E3 ligands for certain diseases
In the near future, the classical ubiquitin-mediated protein degradation will remain in focus. It is thus important to extend the repertoire of E3 ligands. For example, HECT-domain enzymes form a thioester bond with ubiquitin, RING domain catalyze the direct transfer of ubiquitin and CDC20 is essential in all cancer types but low expression in majority of normal tissues. This reminds us to extend the repertoire of ligands to E3 ligases with a variety of structural properties and diverse temporal and spatial expression profiles, essentiality in cancer and ligand ability. Exploiting E3 ligases that are essential to the survival of cancer cells is a promising strategy to avoid resistance mechanism.
Novel concepts employing lysosomal degradation pathway
The lysosome is a major degradation pathway utilized by cells to degrade extracellular and intracellular content. Several designs and modes based on lysosome function were reported until know. For example, LYTAC (lysosome targeting chimera) utilizes a glycan tag to mark an extracellular protein of interest (POI) for intracellular lysosomal degradation following receptor-mediated internalization. AUTAC (autophagy-targeting chimera) binds to the POI and add a degradation tag mimicking S-guanylation, a post-translational modification that triggers K63 polyubiquitination (Ub) of the POI. ATTEC (autophagosome-tethering compound) interact with both the POI and LC3, tethering the POI to the phagophores or autophagosomes for subsequent autophagic degradation.
PART 4
A quick recap
A Broad New Modality Employing Diverse Bifunctional Molecules
In summary, with growing understanding of intracellular and extracellular degradation pathway and advanced techniques, we have discussed the application of bifunctional molecules in protein degradation. This imaginative new modality may enable new routes to functional selectivity and make targeted protein degradation as a creative therapeutic strategy in drug discovery.
Through many years of efforts and development, WuXi AppTec HitS has built an integrated platform that enables discovery of diverse bifunctional molecules:
- Novel chemical modalities and screening methods like DEL and fragment screening
- State-of-the art biophysical methods for ligand characterization
- Structural biology tools to dissect MoA
- Computational design tools for linker design and design of bifunctional molecules
- In vitro functional assays
In addition to HitS Unit, WuXi AppTec also owns the Chemical Services Unit (CSU), which offers a library of tool molecules and customized synthesis services, and HD Biosciences (HDB), which offers state-of-the-art biophysical and biochemical methods such as AlphaScreen, NanoBRET and NanoBiT, which make WuXi AppTec an ideal partner for protein degraders, such as PROTAC developers. For more information on these platforms, please contact:
- WuXi AppTec HitS: HitS_service@wuxiapptec.com
- HD Biosciences (HDB): yang_xiaojie2401@wuxiapptec.com
- Chemical Service Unit (CSU): yu_hao0301@wuxiapptec.com
To achieve the vision that “every drug can be made and every disease can be treated.” WuXi AppTec is committed to enabling companies in the pharmaceutical, biotech and medical device industries worldwide to advance discoveries and deliver groundbreaking treatments to patients.
PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license
Related Content
STAT proteins are key mediators of cellular immunity, proliferation, apoptosis, and differentiation. STATs are known to play a role in...
VIEW RESOURCEAt the Covalent Drug Discovery 2023 Summit and Fragments 2024 Conference, we presented a case study on Bruton’s tyrosine kinase (BTK)...
VIEW RESOURCE
