Establishment of drug induced resistant tumor models enable the development of next generation anticancer therapeutics
AACR Annual meeting 2021
Ting Ni, Zhixiang Zhang, Xuzhen Tang, Hui Qi, Jingying Zhang, Ke Mao, Wenting Shi, Qingyang Gu, Qunsheng Ji
WuXi AppTec, Suzhou, China
Abstract
Recent advances in cancer biology and diagnosis are providing more targeted approaches to treat cancers. Therapy targeting the specific oncogenic driver could inhibit tumor progression and helps to a favorable prognosis in clinical practice. Recently, the discovery of driver mutations in various cancers such as EGFR, ER, CDK, c-MET, TRK and BTK provides a personalized targeted treatment. However, most patients who initially benefit a lot from targeted therapies eventually become resistant to treatments. Acquired resistance limits the long-term efficacy. The commonly acquired resistant mechanisms include target gene mutation, alternative pathway activation, histological or phenotypic transformation and so on. Although a variety of mechanisms of acquired resistance have been reported, optimal treatment for acquired resistance is not yet clearly defined.
To mimic the clinical resistance after long-term treatment, we established a panel of in vivo induced drug resistant CDX and PDX models by continuous dosing of targeted drugs to mice, covering a series of first-line targeted drugs such as Tamoxifen, Olaparib, BMN-673, Ibrutinib, Crizotinib, Erlotinib, Palbociclib, Capmatinib, Entrectinib, Cabozantinib, Sorafenib and Irinotecan. Resistant models were derived from sensitive models harboring specific oncogenic driver. Regrowth tumors were continually treated with targeted drugs and continuously implanted to the next passage in mice until a stable drug resistance phenotype occurred. To explore the resistant mechanisms, we performed WES and RNA-seq analysis in the gene expression levels, and western blotting and IHC staining in the protein expression levels. In summary, these drug induced resistant tumor models provide an opportunity to evaluate the next-generation anticancer therapeutics to overcome resistance in tumors.
Method
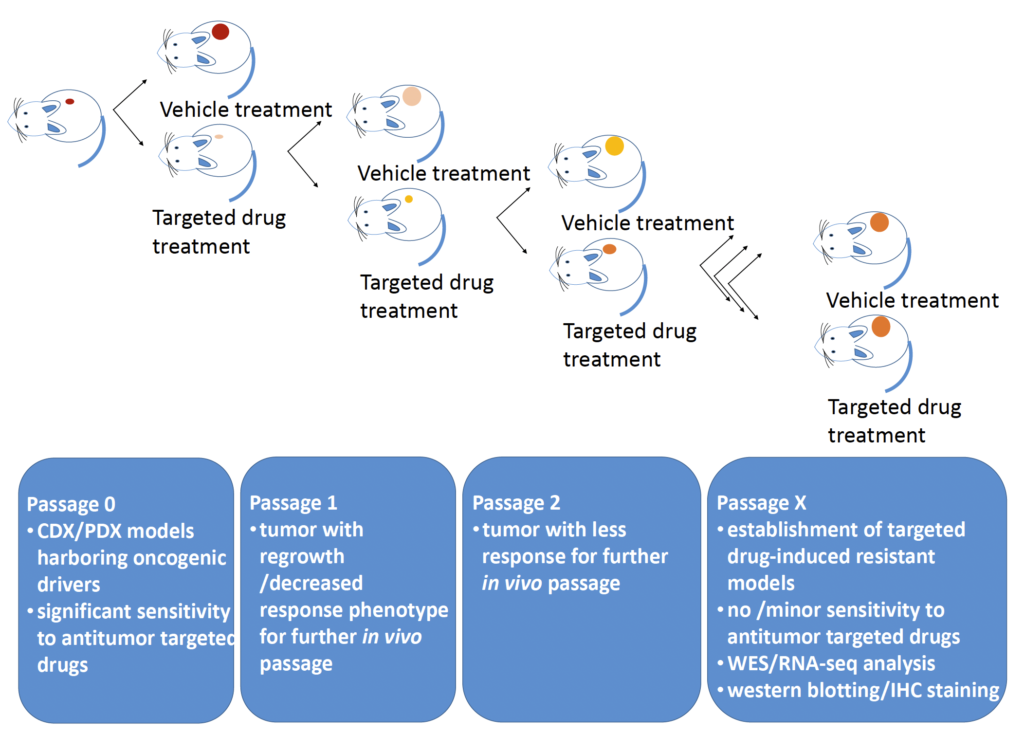
Fig 1. Generation of targeted drug-induced resistant CDX/PDX models
Results
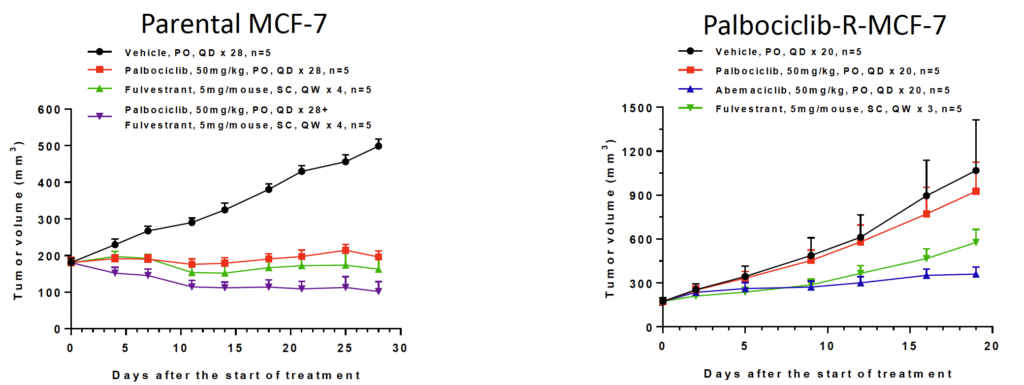
Fig 2. In vivo induced Palbociclib resistant MCF-7 model
Palbociclib, a CDK4/6 inhibitor, has been approved for advanced or metastatic HR+, HER2- breast cancer. Palbociclib-R-MCF-7 model is resistant to Palbociclib, while is sensitive to Abemaciclib (CDK4/6 inhibitor) and Fulvestrant (estrogen receptor antagonist).
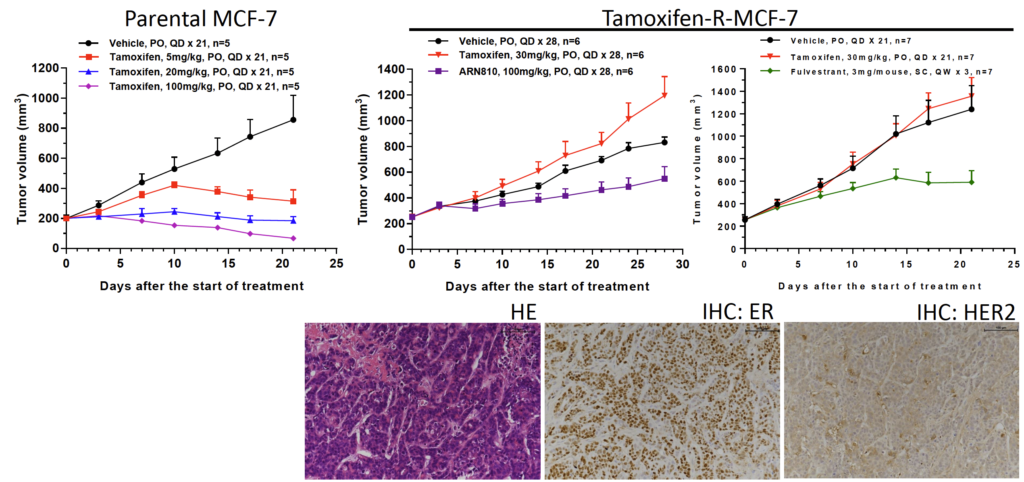
Fig 3. In vivo induced Tamoxifen resistant MCF-7 model
Tamoxifen is a selective estrogen receptor modulator to treat breast cancer patients diagnosed with ER+ after surgery, radiation and/or chemotherapy. In Tamoxifen-R-MCF-7 model, Fulvestrant and ARN810 (estrogen receptor degrader) produce moderate antitumor activities.
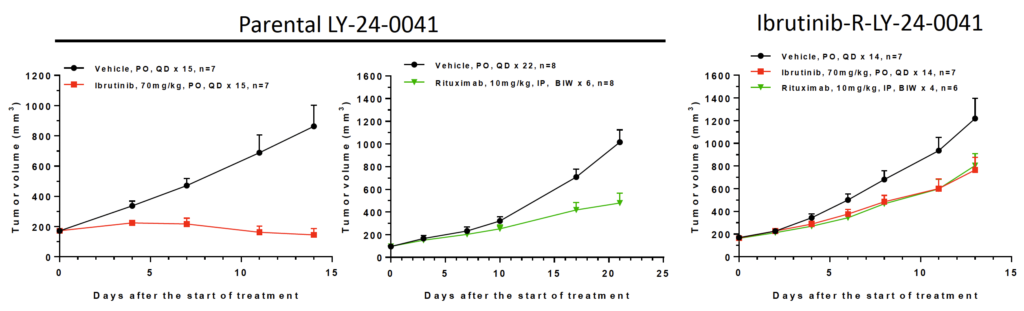
Fig 4. In vivo induced Ibrutinib resistant LY-24-0041 model
Ibrutinib is an orally available, selective irreversible inhibitor of BTK that covalently binds to Cys481. Parental LY-24-0041 PDX model is originally derived from a lymphoma patient and is sensitive to Ibrutinib. Ibrutinib-R-LY-24-0041 model is resistant to Ibrutinib, while has the similar sensitivity to Rituximab (anti-CD20) with the parental model.

Fig 5. In vivo induced Olaparib resistant MDA-MB-436 model
Olaparib is a PARP inhibitor which induces synthetic lethality in BRCA1/2 deficient tumor cells. Olaparib produces a significant antitumor efficacy in beast cancer cell line MDA-MB-436 harboring BRCA1 mutation, while a minor efficacy in Olaparib resistant model.
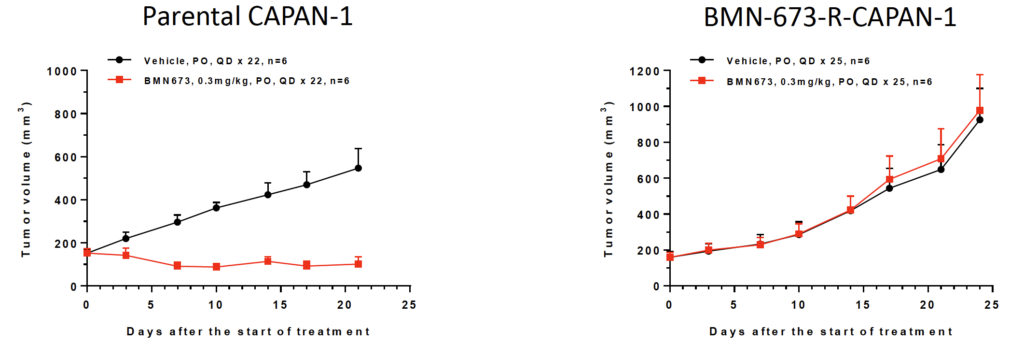
Fig 6. In vivo induced BMN-673 resistant CAPAN-1 model
BMN-673 has been approved as a PARP inhibitor for patients with deleterious or suspected germline BRCA-mutated, HER2-negative locally advanced or metastatic breast cancer. CAPAN-1 is a pancreatic cancer cell line with mutated BRCA2. In BMN-673-R-CAPAN-1 model, BMN-673 produces no antitumor efficacy.
Summary
- We have successfully established a series of in vivo induced drug resistant CDX and PDX models by continuous dosing of targeted drugs.
- In these resistant models, we also validated the efficacy of other drugs in the same or different pathways which are potent in the parental models.
- Furthermore, we performed WES and RNA-seq analysis in the parental and resistant models to explore the mechanisms.
Reference
- Mónica lvarez-Fernández, Malumbres M . Mechanisms of Sensitivity and Resistance to CDK4/6 Inhibition[J]. Cancer Cell, 2020, 37(4):514-529.
- Lai A , Kahraman M , Govek S , et al. Identification of GDC-0810 (ARN-810), an Orally Bioavailable Selective Estrogen Receptor Degrader (SERD) that Demonstrates Robust Activity in Tamoxifen-Resistant Breast Cancer Xenografts.[J]. Journal of Medicinal Chemistry, 2015, 58(12):4888.
- Herrera A F , Jacobsen E D . Ibrutinib for the treatment of mantle cell lymphoma[J]. Clinical Cancer Research, 2014, 20(21):5365-5371.
- Rouleau, Michèle, Patel A , Hendzel M J , et al. PARP inhibition: PARP1 and beyond[J]. Nature Reviews Cancer, 2010, 10(4):293-301.

2021 AACR Drug induced resistant tumor models_2958_Ting Ni_F
Related Content
Rat tumor models are a better mimic of human pathology compared to mice. The tumor stroma component of rats is...
VIEW RESOURCEThe efficacy of CAR-T therapy is substantially influenced by the tumor microenvironment. This is particularly evident when PD-L1 is activated...
VIEW RESOURCE
